Important Dates
| Release of data, code, and metrics for training | Nov. 15, 2021 |
| Release of examples for submission files | Jan. 21, 2022 |
| Release of data and metrics for testing | Jan. 31, 2022 |
| Challenge workshop website goes live | Feb. 01, 2022 |
| Submission deadline of results and papers | Feb. 28, 2022 |
| Response from reviewers | Mar. 21, 2022 |
| Registration deadline and BRIGHT Workshop | Mar. 27, 2022 |
| Camera Ready Version of the paper | Apr. 03, 2022 |
| Post-workshop leaderboard release | Apr. 04, 2022 |
| Submission deadline for manuscripts | Apr. 21, 2022 |
| Publication of challenge outcomes | Oct. 01, 2022 |
Like the BRIGHT Challenge?
Try the KNIGHT challenge for kidney images.
Evaluation
The methods developed by the participants will be evaluated on an external WSI test set for which the ground-truth will be hidden. Participants will be requested to submit a .csv file containing row-wise tuples for all the WSIs in the test set. The tuples must comply to the following scheme:
[ image name, predicted label, class 1 score, ..., class C score ]
where \mathcal{|C|} =\{3, 6\} for Task 1 and Task 2, respectively, and the scores represent the probabilities \in [0,1] of an image to belong to the different classes.
For both the classification tasks, Averaged F1-score will be used to measure performance, given as,
\text{Avg. F1-score}=\frac{1}{\mathcal{C}} \sum_{c \in \mathcal{C}} \, \frac{TP_{c}}{TP_{c} + 1/2 (FP_{c} + FN_{c})}
where, TP_c=True Positive, TN_c=True Negative, FP_c=False Positive, and FN_c=False Negative for class c \in \mathcal{|C|}, and \mathcal{|C|} is the number of classes in a task.
Avg. F1-score will be used to rank the methods, where a higher value of the metric conveys better classification performance.
Submissions that contain missing results will not be processed.
We will also compute the Brier Skill Score (BSS), which is a statistical metric that focuses on the positive class, i.e. the minority class for an imbalanced classification task. BSS is based on the Brier Score (BS), which is calculated as, BS=(1/N) \sum_{i}^{N}(\hat{y}_{i} - y_{i})^{2}
BS=\frac{1}{N}\sum_{i}^{N}(\hat{y}_{i} - y_{i})^{2}
where, \hat{y}=expected probabilities for the positive class, and
y=predicted probabilities. Then, BSS= 1 - (Bs /NSC).
BSS is computed as,
BSS = 1 - (BS / NSC)
BSS scales the BS against the score from a no skill classifier (NSC), with BSS=0, <0 and =1 representing no skill, worse than no skill, and perfect skill, respectively. BSS will be used as an additional evaluation to break ties among methods resulting in comparable Avg. F1-scores.
The testing dataset will be released for public use after the completion of the challenge along with the rest of the BRACS dataset and marked accordingly, following the aforementioned structure.
We will organize a workshop where the summary of the challenge including all results, learned lessons, and generated insights will be presented. We aim to publish all participating methods. Further, the top teams will be included in a journal publication. We also explore, beyond scientific, also financial or in-kind rewards for the top 3 performers. The schedule of the organization is outlined in the Tentative schedule on the left.
We encourage all participants to create multi-institutional teams and make their methods publicly available to support transparency, reproducibility, and to be leveraged by the clinical community. Based on the submitted methods and results we may encourage some teams to potentially collaborate to achieve better results by combining methods. We will publish the winning methods and their results in a journal with the challenge description and goals and the overall evaluation performed in the challenge. We will pay special attention to the usability and reproducibility of the methods to address the critical task of classifying breast tumor lesions. In that regard, to declare the winning method or methods, the top performing submitted models will be requested to be made available through a Docker image and their final results must be reproduced independently through the proposed evaluation platform.
The evaluation script is implemented using FuseMedML , an open-source PyTorch-based framework designed to enhance collaboration, facilitate deep learning R&D and share expertise in medical imaging and multimodal data. The script will be published and can be used to evaluate models through the training process.
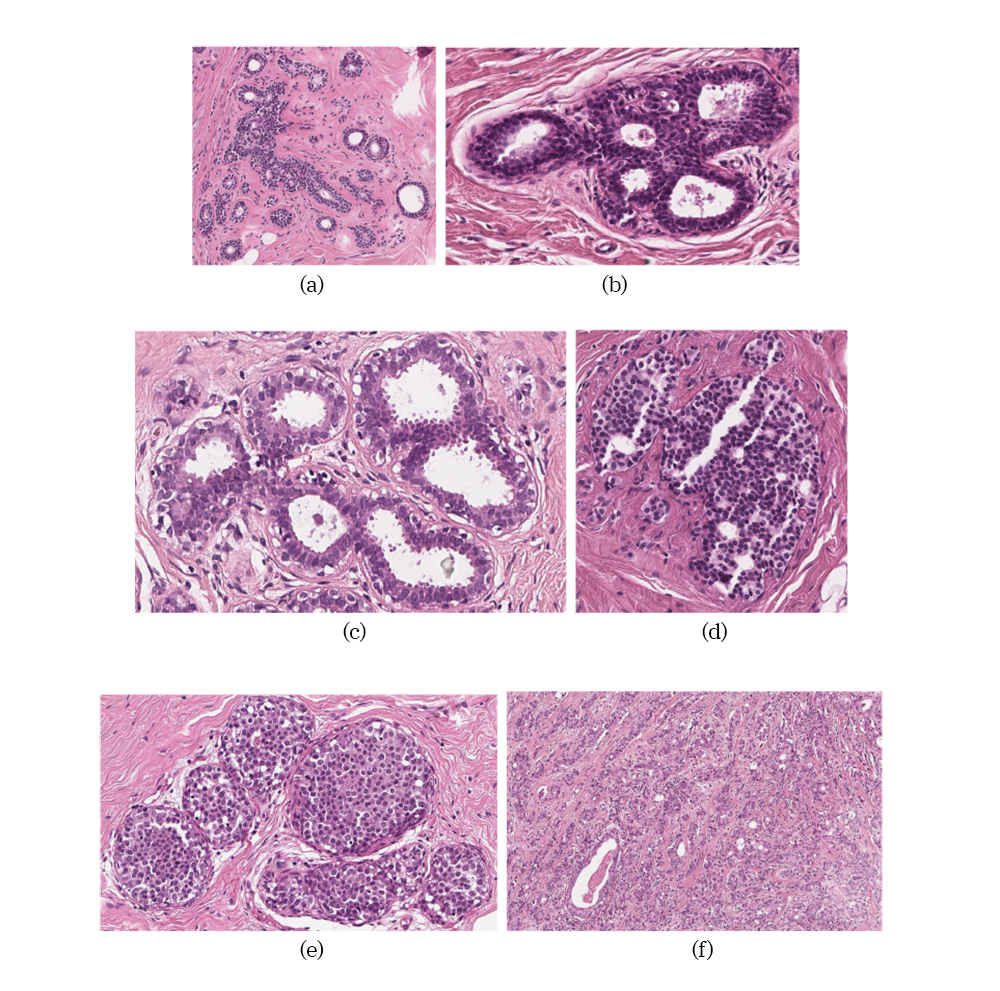
Examples of different tissue samples: a) Pathological Benign (PB), b) Usual Ductal Hyperplasia (UDH), c) Flat Epithelial Atypia (FEA), d) Atypical Ductal Hyperplasia (ADH), e) Carcinoma in Situ (DCIS) and f) Invasive Carcinoma (IC).
- Registration
- Submission (TBD)
- Contact the Organizers
- Leaderboard (TBD)
